The piRNA Pathway - A Small RNA Based Genome Immune System
Throughout the eukaryotic lineage, small RNA silencing pathways safeguard the genome against selfish genetic elements such as transposons. In animals a specialized pathway centered on PIWI proteins and their interacting piRNAs silences transposons within gonads. Genetic and bioinformatics studies have uncovered the fascinating conceptual framework of this pathway that is conserved from invertebrates to mammals. Our group systematically dissects the piRNA pathway regarding its molecular architecture as well as its biological functions in Drosophila.
The importance of silencing selfish genetic elements
Eukaryotic genomes are densely populated by selfish genetic elements. Among those, the most widespread members belong to the class of mobile elements, called transposons. In humans, for example, close to 50% of the entire genome is comprised of transposons and their sequence remnants. The remarkable success of these “genome parasites” rests on their ability to multiply within the genome by transposition to new sites. This leads to widespread defects ranging from insertional mutagenesis to ectopic chromosomal recombination, ultimately resulting in the reduced long-term fitness of the host. The acute threat posed by transposable elements has triggered the evolution of powerful defense-systems in eukaryotes. Though early genetic studies pointed to the existence of such host defense-systems, their molecular nature remained mysterious for a long time. After the discovery of RNA interference (RNAi) in 1998, however, it has become increasingly evident that this regulatory mechanism is at the root of the eukaryotic answer to the transposon challenge.
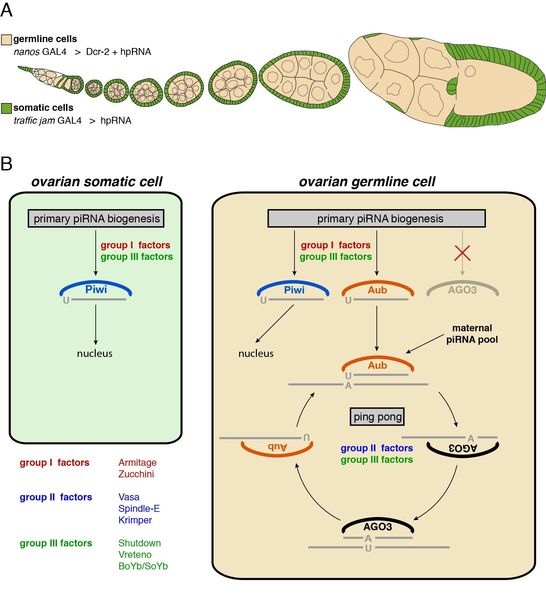
Figure 1: Scheme illustrating the piRNA pathway acting in the Drosophila ovary. (A) This cartoon depicts an ovariole, one of the functional units of an ovary. The two major cell types (somatic support cells and germline cells) are shown in green and beige. (B) Schematic summary of the piRNA pathway architecture in somatic (left) and germline cells (right). Depicted is the wiring of the three PIWI family proteins Piwi, Aubergine and AGO3 into piRNA biogenesis processes. Also indicated are genetically identified piRNA biogenesis factors that act at the various indicated steps.
The piRNA pathway - a small RNA based genome immune system
The piRNA pathway is an evolutionarily conserved small RNA silencing pathway acting in the animal germline. It is the key genome surveillance system that suppresses the activity of transposons. Over the last 5-10 years a conceptual framework for this pathway emerged: The genome stores transposon sequences in defined heterochromatic loci called piRNA clusters. These provide the RNA substrates for the biogenesis of 23-29 nt long piRNAs. Within germline cells, an intricate amplification cycle steers piRNA production predominantly to those cluster regions that are complementary to transposons being active at a given time. Finally, piRNAs guide a protein complex centered on PIWI-proteins to complementary transposon RNAs in the cell, leading to their silencing (Figure 1A, B). In stark contrast to other RNAi pathways, the mechanistic framework of the piRNA pathway is largely unknown. We are for example only at the very beginning to understand processes such as piRNA biogenesis or PIWI mediated silencing. Moreover, the spectrum of biological processes impacted by the piRNA pathway is only poorly understood. piRNAs are not only derived from transposon sequences but also from various other genomic repeats that are enriched at telomeres or heterochromatin. The presence of repetitive sequences throughout the genome is probably being used in order to control vital aspects of chromosome biology. It would not be too surprising if the piRNA pathway were also a key player in this more mutual relationship between genome and transposons. To study this fascinating genome surveillance system, we use Drosophila melanogaster as a model system. For most projects, we combine genetics, biochemistry, cell biology and bioinformatics in often-unique ways.
The main areas of our interest are:
-
Identifying and characterizing novel piRNA pathway members: We have established robust RNAi conditions for both, the somatic ovarian cells where a simplified piRNA pathway is active, but also for germline cells, where many piRNA pathway factors are acting specifically (Figure 1, 2). Using these in vivo RNAi systems we performed genome wide screens and identified several novel piRNA pathway genes in Drosophila. Their genetic and molecular characterization promises a deeper understanding of essentially all levels of this pathway, from piRNA cluster biology to piRNA biogenesis and to piRNA mediated silencing.
-
Systems level analysis of gene/transposon expression in wildtype and piRNA pathway mutants: Till today, no systematic analysis on transposon activity and transposition frequency and patterns has been conducted in flies lacking the piRNA pathway. Using our established RNAi conditions we will probe the genome wide consequences of deficiencies in the somatic and germline piRNA pathways. We are taking advantage of deep sequencing technologies coupled to bioinformatics to obtain novel insight into these questions.
-
Understanding the biology of piRNA clusters: piRNA clusters are at the heart of the pathway as they serve as sequence repositories for transposons. They are typically located at telomeres or at the border between euchromatin and heterochromatin. Their transcripts are believed to traverse large (up to several hundreds of kb) heterochromatic regions. We are interested in transcription, specification, export and processing of piRNA clusters or their transcripts. Ultimately, we want to understand how the cell discriminates cluster transcripts from other RNAs in the cell.
Figure 2: Efficient RNAi approaches for somatic and germline cells in the ovary. Shown are immuno-fluorescence images from single egg chambers stained for the essential piRNA biogenesis factor Armitage. To the left, Armitage expression and localization in wildtype egg chambers is seen in somatic and germline cells. Upon in vivo RNAi in either somatic cells (center) or germline cells (right) Armitage protein is essentially lost in the respective cell type.